See also
This page was generated from examples/example09.ipynb.
Download the Jupyter Notebook for this section: example09.ipynb. View in nbviewer.
MinDE System with Spatiocyte Simulator¶
[1]:
%matplotlib inline
from ecell4.prelude import *
Declaring Species and ReactionRules:
[2]:
with species_attributes():
cytoplasm | {'radius': 1e-8, 'D': 0, 'dimension': 3}
MinDatp | MinDadp | {'radius': 1e-8, 'D': 16e-12, 'location': 'cytoplasm', 'dimension': 3}
MinEE_C | {'radius': 1e-8, 'D': 10e-12, 'location': 'cytoplasm', 'dimension': 3}
membrane | {'radius': 1e-8, 'D': 0, 'location': 'cytoplasm', 'dimension': 2}
MinD | MinEE_M | MinDEE | MinDEED | {'radius': 1e-8, 'D': 0.02e-12, 'location': 'membrane', 'dimension': 2}
with reaction_rules():
membrane + MinDatp > MinD | 2.2e-8
MinD + MinDatp > MinD + MinD | 3e-20
MinD + MinEE_C > MinDEE | 5e-19
MinDEE > MinEE_M + MinDadp | 1
MinDadp > MinDatp | 5
MinDEE + MinD > MinDEED | 5e-15
MinDEED > MinDEE + MinDadp | 1
MinEE_M > MinEE_C | 0.83
m = get_model()
Make a World.
[3]:
f = spatiocyte.Factory(1e-8)
w = f.world(Real3(4.6e-6, 1.1e-6, 1.1e-6))
w.bind_to(m)
Make a Structures.
[4]:
rod = Rod(3.5e-6, 0.51e-6, w.edge_lengths() * 0.5)
w.add_structure(Species('cytoplasm'), rod)
w.add_structure(Species('membrane'), rod.surface())
[4]:
47496
Throw-in molecules.
[5]:
w.add_molecules(Species('MinDadp'), 1300)
w.add_molecules(Species('MinDEE'), 700)
[5]:
True
Run a simulation for 240 seconds.
[6]:
sim = f.simulator(w, m)
[7]:
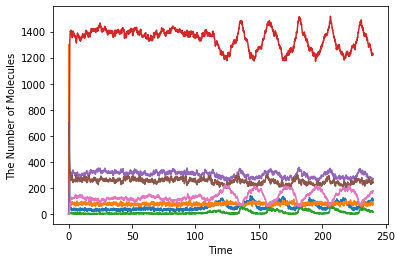
obs1 = FixedIntervalNumberObserver(0.1, ('MinDatp', 'MinDadp', 'MinEE_C', 'MinD', 'MinEE_M', 'MinDEE', 'MinDEED'))
[8]:
obs2 = FixedIntervalHDF5Observer(1.0, "minde%03d.h5")
[9]:
duration = 240
[10]:
sim.run(duration, (obs1, obs2))
[11]:
show(obs1)
[12]:
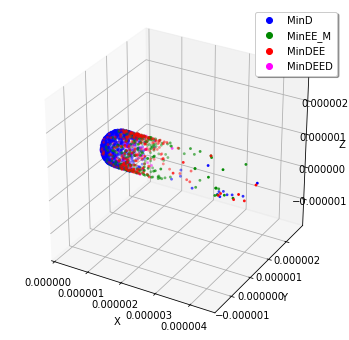
plotting.plot_movie([spatiocyte.SpatiocyteWorld("minde%03d.h5" % i) for i in range(obs2.num_steps())], species_list=('MinD', 'MinEE_M', 'MinDEE', 'MinDEED'))
[13]:
plotting.plot_world(spatiocyte.SpatiocyteWorld("minde240.h5"), species_list=('MinD', 'MinEE_M', 'MinDEE', 'MinDEED'))