See also
This page was generated from top.ipynb.
Download the Jupyter Notebook for this section: top.ipynb. View in nbviewer.
Welcome to E-Cell4 in Jupyter Notebook!¶
Quick Start¶
Binding and unbinding reactions¶
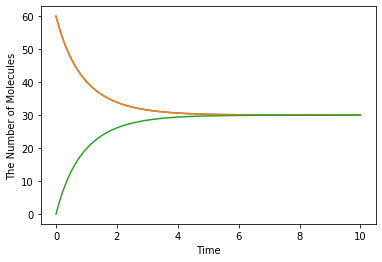
Here is an extremely simple example with a reversible binding reaction:
[1]:
%matplotlib inline
from ecell4.prelude import *
with reaction_rules():
A + B == C | (0.01, 0.3)
run_simulation(10, {'A': 60, 'B': 60})
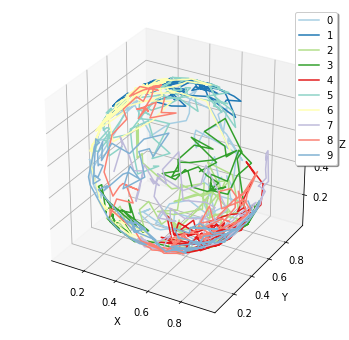
Diffusion on a spherical surface¶
[2]:
%matplotlib inline
from ecell4.prelude import *
with species_attributes():
M | {'dimension': 2}
A | {'D': 1.0, 'location': 'M'}
surface = Sphere(ones() * 0.5, 0.49).surface()
obs = FixedIntervalTrajectoryObserver(4e-3)
run_simulation(
0.4, y0={'A': 10}, structures={'M': surface},
solver='spatiocyte', observers=obs)
show(obs)
Plotly¶
Interactive visualizations by Plotly are also available. This part might not works on Google Colab.
[3]:
%matplotlib inline
from ecell4.prelude import *
[4]:
with reaction_rules():
A + B == C | (0.01, 0.3)
ret = run_simulation(10, {'A': 60, 'B': 60})
[5]:
ret.plot(backend='plotly')
[6]:
with species_attributes():
M | {'dimension': 2}
A | {'D': 1.0, 'location': 'M'}
ret = run_simulation(
0.4,
y0={'A': 10},
structures={'M': Sphere(ones() * 0.5, 0.49).surface()},
solver='spatiocyte',
observers=FixedIntervalTrajectoryObserver(1e-3))
[7]:
plotting.plot_world(ret.world, species_list=['M', 'A'], backend='plotly')
[8]:
plotting.plot_trajectory(ret.observers[1], backend='plotly')