See also
This page was generated from tests/Birth_Death.ipynb.
Download the Jupyter Notebook for this section: Birth_Death.ipynb. View in nbviewer.
Birth-Death¶
This is for an integrated test of E-Cell4. Here, we test a simple birth-death process in volume.
[1]:
%matplotlib inline
from ecell4.prelude import *
Parameters are given as follows. D and radius mean a diffusion constant and a radius of molecules, respectively. Dimensions of length and time are assumed to be micro-meter and second.
[2]:
D = 1 # 0.01
radius = 0.005
[3]:
N = 20 # a number of samples
[4]:
y0 = {} # {'A': 60}
duration = 3
V = 8
Make a model for all algorithms. No birth reaction with more than one product is accepted.
[5]:
with species_attributes():
A | {'radius': radius, 'D': D}
with reaction_rules():
~A > A | 45.0
A > ~A | 1.5
m = get_model()
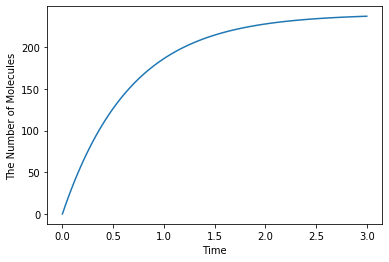
Save a result with ode as obs, and plot it:
[6]:
ret1 = run_simulation(duration, y0=y0, volume=V, model=m)
ret1
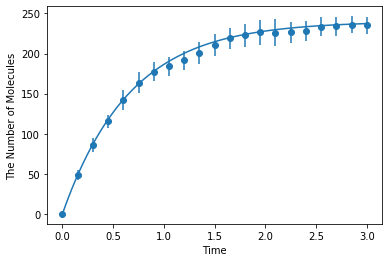
Simulating with gillespie (Bars represent standard error of the mean):
[7]:
ret2 = ensemble_simulations(duration, ndiv=20, y0=y0, volume=V, model=m, solver='gillespie', repeat=N)
ret2.plot('o', ret1, '-')
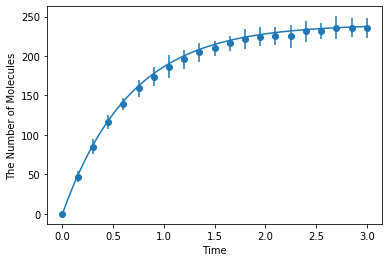
Simulating with meso:
[8]:
ret2 = ensemble_simulations(
duration, ndiv=20, y0=y0, volume=V, model=m, solver=('meso', Integer3(3, 3, 3), 0.25), repeat=N)
ret2.plot('o', ret1, '-')
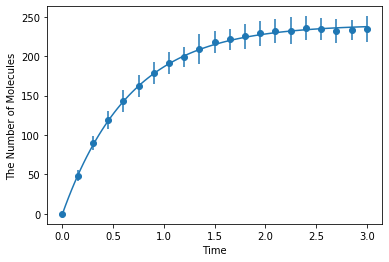
Simulating with spatiocyte:
[9]:
ret2 = ensemble_simulations(
duration, ndiv=20, y0=y0, volume=V, model=m, solver=('spatiocyte', radius), repeat=N)
ret2.plot('o', ret1, '-')
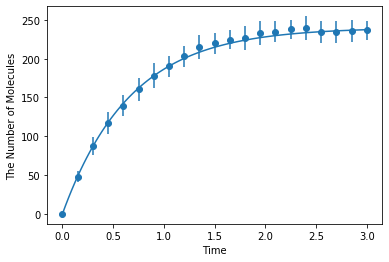
Simulating with egfrd:
[10]:
ret2 = ensemble_simulations(
duration, ndiv=20, y0=y0, volume=V, model=m, solver=('egfrd', Integer3(8, 8, 8)), repeat=N)
ret2.plot('o', ret1, '-')
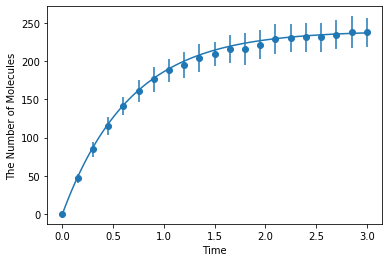
Simulating with bd:
[11]:
ret2 = ensemble_simulations(
duration, ndiv=20, y0=y0, volume=V, model=m, solver=('bd', Integer3(8, 8, 8), 0.1), repeat=N)
ret2.plot('o', ret1, '-')